
We recommend using the plugins for Flye or Minimap2 for assembly and mapping of Oxford Nanopore and PacBio data, rather than using the Geneious assembler.
Depending on the data quality, you may be able to use these with map to reference, but are unlikely to be able to successfully de novo assemble them. PacBio CLR and Oxford Nanopore data are more problematic for Geneious due to the very high error rates.

The Geneious assembler works well with Sanger, Illumina, Ion Torrent, 454, and PacBio CCS data. This can also be set during the file import process if you are importing fastq files. To ensure the optimal parameters are used for your dataset you can set the sequencing platform under Sequence → Set Read Technology. Because different sequencing platforms have different error profiles and rates, they may require different assembly parameters. The Geneious assembler can handle data from Sanger and high-throughput (NGS) sequence platforms. The first approach is usually applied to genomes that have not been characterised yet, while the second one usually focuses on identifying differences from a well-characterised reference sequence. De novo assembly focuses on the reconstruction of the original sequence by aligning and merging shorter reads, while map to reference consists of mapping reads to a reference sequence.
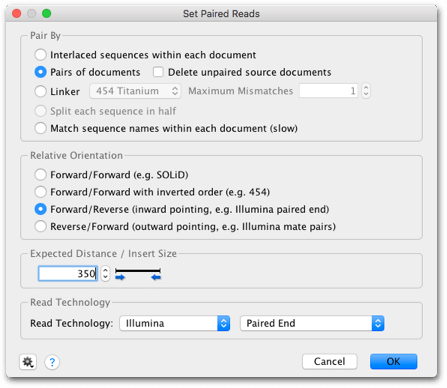
Sequence assembly can refer either to de novo assembly or map to reference. Where positional information such as paired-end and mate-pair data is available, contigs can be joined into longer sequences called scaffolds. The assembly essentially appears as a multiple sequence alignment of reads (called the contig document) and the consensus sequence of the contig can be used for the reconstruction of the original sequence. Assembly is normally used to align and merge overlapping fragments of a DNA sequence (typically produced from Sanger or next-generation sequencing (NGS) sequence platforms) to reconstruct the original sequence.


 0 kommentar(er)
0 kommentar(er)
